QuickSwitch™ Custom Tetramer Kits
Fast, High-Quality Generation of New Tetramer Specificities
Rapid, high-quality creation of custom Class I and Class II MHC tetramers in your own lab is now a reality! Create new specificity tetramers in just a few hours with QuickSwitch™, a proprietary technology for exchanging peptides on an MHC tetramer. With this kit, you will receive MHC allele with an irrelevant peptide bound. The addition of a peptide exchange factor, and your peptide of interest, results in the creation of your custom MHC tetramer ready to use- all in one day. The QuickSwitch™ Quant Tetramer Kit can quantify the exchange with the new peptide, thereby gathering binding affinity information for the MHC/peptide complex. QuickSwitch™ can be used to generate custom MHC tetramers, or to screen numerous peptides. This kit can be used for purposes such as epitope discovery, neoantigen vaccine research, verification of T cell staining using the new peptide specific tetramer, and more.
Functional screening of peptides for MHC class I binding is essential for vaccine design and immune monitoring. The QuickSwitch™ assay kit allows discrimination of MHC binding from non-binding peptides. This is particularly essential for the screening of immunogenic peptides from infectious agents or cancer neoantigens. Tetramers resulting from peptide exchange with selected peptides can then be used for immune monitoring. QuickSwitch™ kits are optimized for up to 10 peptide exchanges and multiple tests per resulting tetramer.
View All QuickSwitch™ Products
QuickSwitch™ Class I Custom Tetramer Kit
Peptide exchange, quantification, cell staining, and flow cytometry analysis can all be performed in one day!
Steps for custom Class I MHC Tetramer creation*
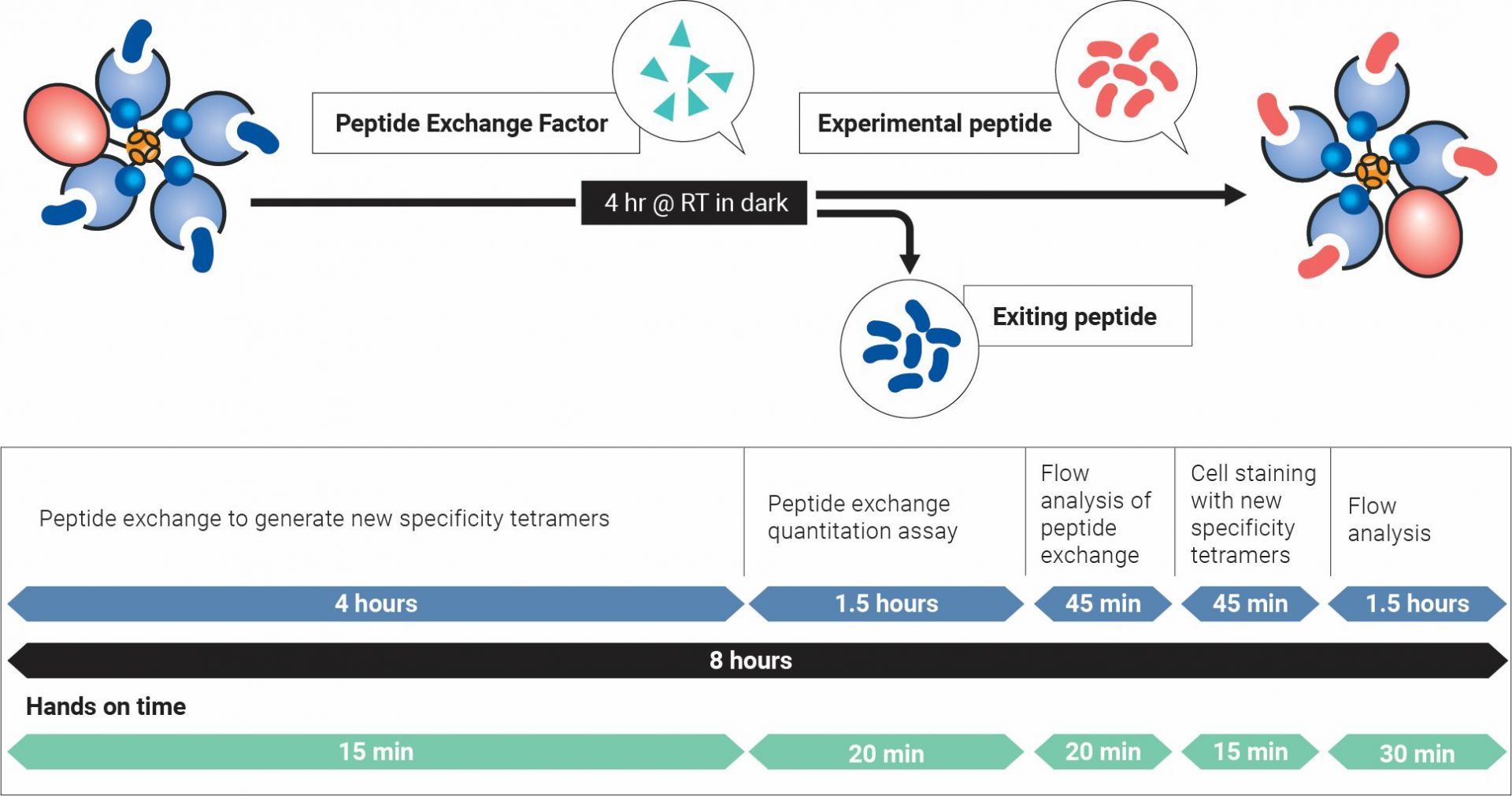
*Total assay time is dependent on allele version of QuickSwitch™ kit.
High Throughput Immunogenic Peptide Discovery and Validation in 90 min.*
- Validate MHC binding peptides from in silico selected list of candidate peptides
- Generate new specificity tetramers for immune monitoring
- Perform functional stability studies for MHC binding peptides
- Compare epitopes to rank better binders and perform epitope mapping
Class I Kits Available
| Product Description | ALLELE | Conjugates Available | Quantification Included |
|---|---|---|---|
| QuickSwitch™ Quant HLA-A*01:01 Tetramer Kit | HLA-A*01:01 | PE, APC, BV421 | Yes |
| QuickSwitch™ Quant HLA-A*02:01 Tetramer Kit | HLA-A*02:01 | PE, APC, BV421 | Yes |
| QuickSwitch™ Quant HLA-A*03:01 Tetramer Kit | HLA-A*03:01 | PE, APC, BV421 | Yes |
| QuickSwitch™ Quant HLA-A*11:01 Tetramer Kit | HLA-A*11:01 | PE, APC, BV421 | Yes |
| QuickSwitch™ Quant HLA-A*24:02 Tetramer Kit | HLA-A*24:02 | PE, APC, BV421 | Yes |
| QuickSwitch™ Quant HLA-B*07:02 Tetramer Kit | HLA-B*07:02 | PE, APC, BV421 | Yes |
| QuickSwitch™ Quant H-2 Kb Tetramer Kit | H-2Kb | PE, APC, BV421 | Yes |
| QuickSwitch™ HLA-A*02:01 Peptide Screening Kit | HLA-A*02:01 | Yes |
QuickSwitch™ Class II Custom Tetramer Kit
Custom Class II MHC Tetramers: Peptide exchange, quantify, cell staining, and FCM analysis all in your own lab!
Steps for custom Class II MHC Tetramer creation
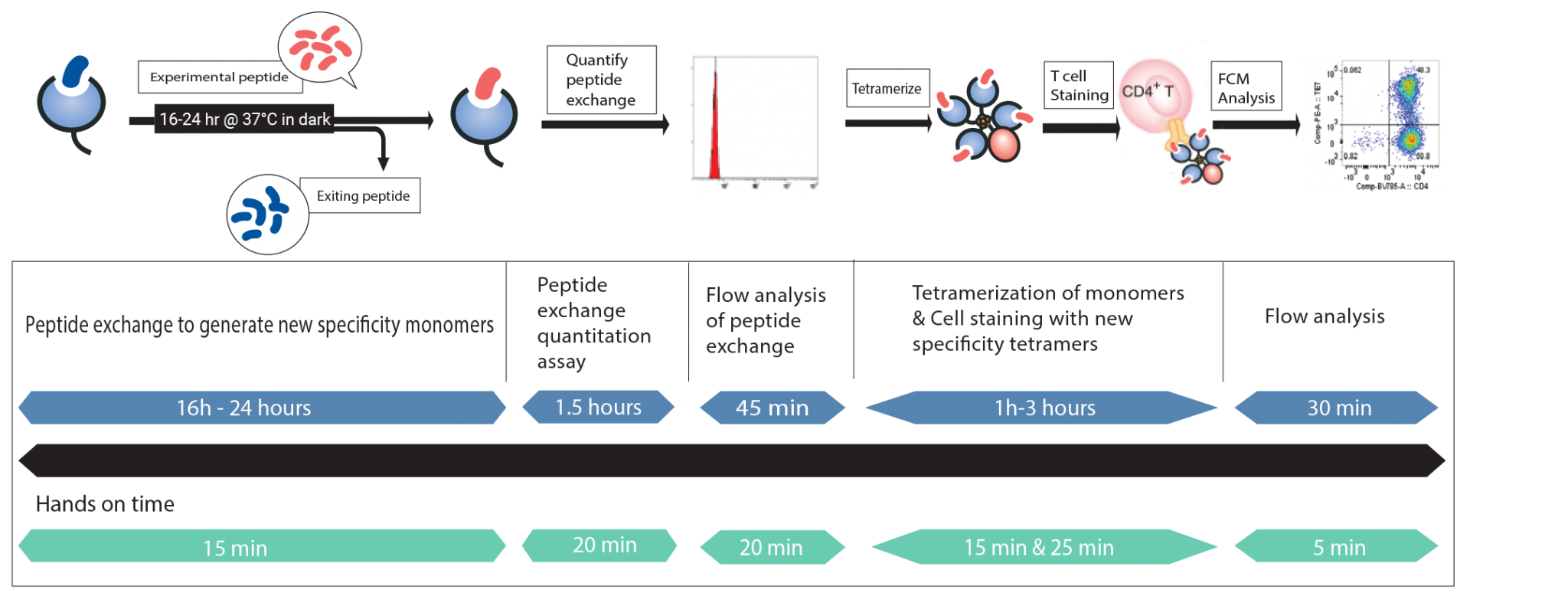
*Total assay time is dependent on allele version of QuickSwitch™ kit.
Class II Kits Available
| Product Description | ALLELE | Conjugates Available | Quantification Included |
|---|---|---|---|
| QuickSwitch™ Quant HLA-DRB1*01:01 Tetramer Kit | HLA-DRB1*01:01 | PE, APC, BV421 | Yes |
| QuickSwitch™ Quant HLA-DRB1*04:01 Tetramer Kit | HLA-DRB1*04:01 | PE, APC, BV421 | Yes |
| QuickSwitch™ Quant HLA-DRB1*15:01 Tetramer Kit | HLA-DRB1*15:01 | PE, APC, BV421 | Yes |
Additional QuickSwitch™ data
QuickSwitch™ HLA-A*02:01 tetramers detect similar percentages of low and high affinity CMV responses in PBMCs as classically folded tetramers

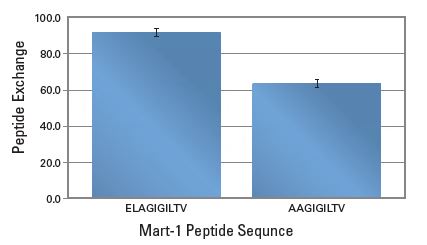
QuickSwitch™ Quant Kit can be used to assess peptide exchange so that you can select peptides with appropriate affinities to make functional tetramers prior to cell staining. In a study where HLA-A*02:01 QuickSwitch™ tetramer was incubated for 4 hours with two Mart-1 related peptides at a final of 20 μM in presence of peptide exchange factor #1, peptide exchange correlated with the theoretical peptide affinity of each peptide towards HLA-A*02:01.
QuickSwitch™ Quant Kit components:
- Tetramer with an irrelevant exchangeable peptide in the MHC groove
- A peptide exchange factor for catalyzing the peptide exchange reaction
- A high affinity MHC-binding reference peptide used as peptide exchange positive control
- FITC conjugated antibody specific of the exiting peptide
- Magnetic beads conjugated with anti MHC antibody for tetramer capture
- An Assay Buffer for diluting reagents and washing steps
H2-Kb Peptide-exchanged tetramers perform similarly to classically folded tetramers
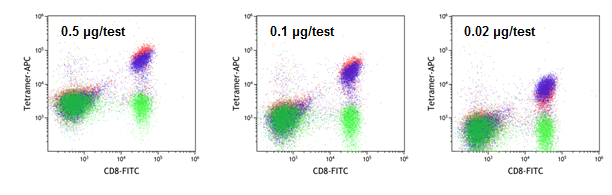
H-2 Kb TRP2 used a negative control (#TB-5004-2; green), classically folded H-2 Kb OVA (#TB-5001-2; blue), and H-2 Kb QuickSwitch™ OVA (red) tetramer staining.
QuickSwitch™ HLA-A*24:02 tetramers perform similarly to classically folded tetramers
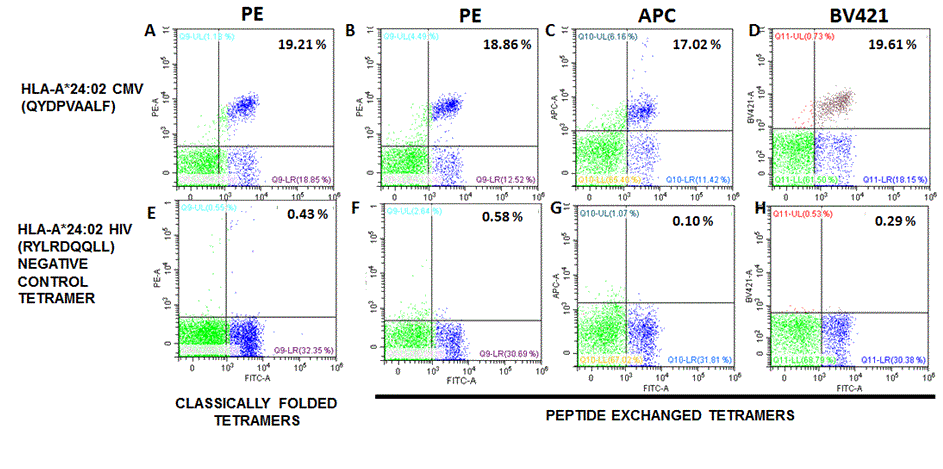
Data show CD3+ PBMCs stained with classically folded tetramers (A,E) or with QuickSwitch™ tetramers obtained by peptide exchange with the HLA-A*24:02 CMV peptide (B,C,D) or the HIV negative control peptide (F,G,H). 2 x 105 cells in 50 μL PBS-BSA-NaN3 buffer were stained for 30 min at RT with 1 μL of anti CD3-PC5.5 mAb (clone OKT3), 1 μL anti CD8-FITC (clone RPA-T8) (MBL) and 0.25 μg tetramer. Cells were fixed with a 0.5% formaldehyde PBS solution. Cells were analyzed on a Cytoflex S flow cytometer (Beckman Coulter). Cell doublets were discriminated using SSC-W/SSC-A gating.
MHC Quant Monomer Kit
Create custom MHC Tetramer from MHC Monomers
MBL International’s MHC Quant Monomer Kit allows you to create your own custom MHC Tetramer from MHC Monomers. You may already know your peptide binds well with the MHC allele or you may want to screen some peptides to determine peptide-MHC binding. This MHC Quant Monomer kit comes with MHC Monomer bound with an irrelevant peptide. Perform peptide exchange on the monomer with your peptides of interest and quantitate the exchange. Then, tetramerize your new MHC Monomers bound with your peptide(s) of interest and add your desired conjugate. The resulting custom MHC Tetramer can be used to detect antigen specific T cells. MBL International’s revolutionary MHC Tetramer technology continues to provide unique products for solutions to advance T cell research.
MBL International continuously updates and adds new alleles to our current offerings. If you do not see your allele of interest, please contact us to inquire for more information.
Learn more about our Related Product Lines
Tetramer Products
MHC tetramers are composed of four monomers, each with a specific peptide.
Peptides Products
MBL International’s stock and custom peptides can be used for stimulation assays or with custom tetramer generation using the QuickSwitch™ custom tetramer kits.
Flow Cytometry Products
These antibodies have high sensitivity and specificity and are validated for numerous applications.
Explore our video library of webinars
Download our free webinars which focus on different life science research topics and applications. All webinars are presented by our scientists at MBL International.
Citations
Aoki, Hiroyasu, et al. "CD8+ T cell memory induced by successive SARS-CoV-2 mRNA vaccinations is characterized by shifts in clonal dominance." Cell Reports (2024).
Wu, Ming, et al. “Spleen‐targeted neoantigen DNA vaccine for personalized immunotherapy of hepatocellular carcinoma.” EMBO Molecular Medicine (2023): e16836.
Qin, Lijuan, et al. “GPC3 and PEG10 peptides associated with placental gp96 elicit specific T cell immunity against hepatocellular carcinoma.” Cancer Immunology, Immunotherapy (2023): 1-18.
Tai, W., Feng, S., Chai, B. et al. An mRNA-based T-cell-inducing antigen strengthens COVID-19 vaccine against SARS-CoV-2 variants. Nat Commun 14, 2962 (2023).
Wang, Wenfeng, et al. “Lymphatic endothelial transcription factor Tbx1 promotes an immunosuppressive microenvironment to facilitate post-myocardial infarction repair.” Immunity (2023).
Xiao, Jingyu, et al. “Engineering In Vitro Organ‐Structured Tumor Model for Evaluating Neoantigen‐Specific T Cell Responses in Hepatocellular Carcinoma.” Advanced Materials Interfaces (2023): 2300155
Shin, Hocheol, Yujin Kim, and Sangyong Jon. “Nanovaccine displaying immunodominant T cell epitopes of fibroblast activation protein is effective against desmoplastic tumors.” ACS nano (2023). 10.1021/acsnano.3c00764
Saotome, K., Dudgeon, D., Colotti, K. et al. Structural analysis of cancer-relevant TCR-CD3 and peptide-MHC complexes by cryoEM. Nat Commun 14, 2401 (2023). doi:10.1038/s41467-023-37532-7
Bruno, Peter M., et al. “High-Throughput, Targeted MHC Class I Immunopeptidomics Using a Functional Genetics Screening Platform.” Nature Biotechnology, 2023, doi:10.1038/s41587-022-01566-x.
Son, Eric T., et al. “Screening Self-Peptides for Recognition by Mouse Alloreactive cd8+ T Cells Using Direct Ex Vivo Multimer Staining.” STAR Protocols, vol. 4, no. 1, 2023, p. 101943., doi:10.1016/j.xpro.2022.101943.
Ogura, Hideki, et al. “DYSFUNCTIONAL SARS-COV-2-M Protein-Specific Cytotoxic T Lymphocytes in Patients Recovering from Severe COVID-19.” Nature Communications, vol. 13, no. 1, 2022, doi:10.1038/s41467-022-34655-1.
Wang, Sizhen, et al. “Expansion of KRAS Hot Spot Mutations Reactive T Cells from Human Pancreatic Tumors Using Autologous T-Cells as the Antigen-Presenting Cells.” 2022, doi:10.21203/rs.3.rs-1944651/v1.
Zhang, Xiaoyan, et al. “Single-Cell Sequencing Reveals cd133+cd44−-Originating Evolution and Novel STEMNESS Related Variants in Human Colorectal Cancer.” EBioMedicine, vol. 82, 2022, p. 104125., doi:10.1016/j.ebiom.2022.104125.
Hikichi, T., Sakamoto, M., Harada, M., Saito, M., Yamane, Y., Tokumura, K., & Nakamura, Y. (2022). Identification of cytotoxic T cells and their T cell receptor sequences targeting COVID-19 using MHC class I-binding peptides. Journal of Human Genetics. https://doi.org/10.1038/s10038-022-01013-4
Dong et al., Proteogenomic characterization identifies clinically relevant subgroups of intrahepatic cholangiocarcinoma, Cancer Cell (2021), https://doi.org/10.1016/j.ccell.2021.12.006
Zhang, SM., Cai, W.L., Liu, X. et al. KDM5B promotes immune evasion by recruiting SETDB1 to silence retroelements. Nature 598,682–687 (2021). Available at: https://doi.org/10.1038/s41586-021-03994-2
Morisaki T, Morisaki T, Kubo M, Onishi H, Hirano T, Morisaki S, Eto M, Monji K, Takeuchi A, Nakagawa S, Tanaka H, Koya N, Umebayashi M, Tsujimura K, Yew PY, Yoshimura S, Kiyotani K, Nakamura Y. Efficacy of Intranodal Neoantigen Peptide-pulsed Dendritic Cell Vaccine Monotherapy in Patients With Advanced Solid Tumors: A Retrospective Analysis. Anticancer Res. 2021 Aug. Available at https://ar.iiarjournals.org/content/41/8/4101.abstract#sec1ps://ar.iiarjournals.org/content/41/8/4101.abstract#sec-1
Sydney X. Lu, Emma de Neef, James D. Thomas, Luis Diaz, Jr., Omar Abdel-Wahab, Robert K. Bradley. (2021) Pharmacologic modulation of RNA splicing enhances anti-tumor immunity. Cell, 12066. Available at: /https://www.sciencedirect.com/science/article/abs/pii/S0092867421006905
Hu, C., Shen, M., Han, X., Chen, Q., Li, L., Chen, S., . . . Jin, A. (2021). Identification of Cross-Reactive CD8 T Cell Receptors with High Functional Avidity to a SARS-CoV-2 Immunodominant Epitope and Its Natural Mutant Variants. Genes & Diseases. Available at: https://www.sciencedirect.com/science/article/pii/S2352304221000842
Han, J., Yu, R., Duan, J., Li, J., Zhao, W., Feng, G., . . . Wang, J. (2021). Weighting tumor-specific TCR repertoires as a classifier to stratify the immunotherapy delivery in non–small cell lung cancers. Science Advances, 7(21). Available at: https://advances.sciencemag.org/content/7/21/eabd6971
Teng Wei, Matthias Leisegang, Ming Xia, Kazuma Kiyotani, Ning Li, Chenquan Zeng, Chunyan Deng, Jinxing Jiang, Makiko Harada, Nishant Agrawal, Liangping Li, Hui Qi, Yusuke Nakamura & Lili Ren (2021) Generation of neoantigen-specific T cells for adoptive cell transfer for treating head and neck squamous cell carcinoma, OncoImmunology, 10:1, Available at: https://www.tandfonline.com/doi/full/10.1080/2162402X.2021.1929726
Poluektov, Y., Daftarian, P., & Delcommenne, M. C. (2020). Assessment of SARS-CoV-2 Specific CD4( ) and CD8 ( ) T Cell Responses Using MHC Class I and II Tetramers. Biorxiv Available at: https://www.biorxiv.org/content/10.1101/2020.07.08.194209v1://www.biorxiv.org/content/10.1101/2020.07.08.194209v1
Son, E. T., Faridi, P., Paul-Heng, M., Leong, M., English, K., Ramarathinam, S. H., . . . Sharland, A. F. (2020). The self-peptide repertoire plays a critical role in transplant tolerance induction. Biorxiv. Available at: https://whttps://www.biorxiv.org/content/10.1101/2020.11.09.359968v2ww.biorxiv.org/content/10.1101/2020.11.09.359968v2
Zhang, Y., Zhang, J., Chen, Y., Luo, B., Yuan, Y., Huang, F., . . . Zhang, H. (2020). The ORF8 Protein of SARS-CoV-2 Mediates Immune Evasion through Potently Downregulating MHC-I. Biorxiv. Available at: https://www.biorxiv.org/content/10.1101/2020.05.24.111823v1/www.biorxiv.org/content/10.1101/2020.05.24.111823v1
Najafabadi, A. H., Zhang, J., Aikins, M. E., Abadi, Z. I., Liao, F., Qin, Y., Moon, J. J. (2020). Cancer Immunotherapy via Targeting Cancer Stem Cells Using Vaccine Nanodiscs. Nano Letters, 20(10), 7783-7792. Available at: https://pubs.acs.org/doi/10.1021/acs.nanolett.0c03414://pubs.acs.org/doi/10.1021/acs.nanolett.0c03414
Song, X., Lu, Z., & Xu, J. (2020). Targeting cluster of differentiation 47 improves the efficacy of anti‑cytotoxic T‑lymphocyte associated protein 4 treatment via antigen presentation enhancement in pancreatic ductal adenocarcinoma. Experimental and Therapeutic Medicine. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7444377/s://www.ncbi.nlm.nih.gov/pmc/articles/PMC7444377/
Lo, W., Parkhurst, M., Robbins, P. F., Tran, E., Lu, Y., Jia, L., . . . Rosenberg, S. A. (2019). Immunologic Recognition of a Shared p53 Mutated Neoantigen in a Patient with Metastatic Colorectal Cancer. Cancer Immunology Research, 7(4), 534-543 Available at: https://cancerimmunolres.aacrjournals.org/content/7/4/534
Dustin, M. L., & Mayya, V. (2019). Faculty Opinions recommendation of T-Scan: A Genome-wide Method for the Systematic Discovery of T Cell Epitopes. Faculty Opinions – Post-Publication Peer Review of the Biomedical Literature. Available at: https://pubmed.ncbi.nlm.nih.gov/31398327/
Smith, C. C., Chai, S., Washington, A. R., Lee, S. J., Landoni, E., Field, K., . . . Vincent, B. G. (2019). Machine-Learning Prediction of Tumor Antigen Immunogenicity in the Selection of Therapeutic Epitopes. Cancer Immunology Research, 7(10), 1591-1604. Available at: https://cancerimmunolres.aacrjournals.org/content/7/10/1591.full-text.pdfps://cancerimmunolres.aacrjournals.org/content/7/10/1591.full-text.pdf
Marc C Delcommenne, Olga Hrytsenko, Cynthia Tram, Genevieve Weir and Marianne M. Stanford. (2017) The QuickSwitch Quant HLA-A*02:01 Tetramer Kit can be used for determining the biological activity of a cancer vaccine. J Immunol, 198 (1 Supplement) 79.27 Available at: https://www.jimmunol.org/content/198/1_Supplement/79.27://www.jimmunol.org/content/198/1_Supplement/79.27