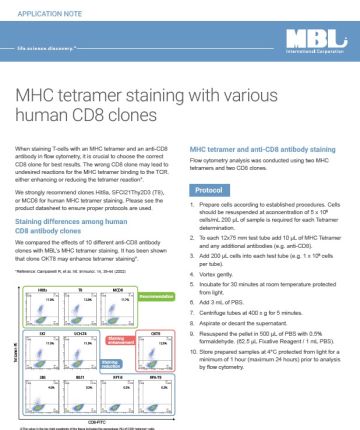
RIP-Assay Kit for microRNA
RIP-Assay Kit for microRNA is optimized to immunochemically isolate endogenous miRNAs, mRNAs and RBPs within mRNP complexes. The kit is designed to isolate cellular miRNAs that being incorporated into the RISC and/or to isolate unique group of miRNAs that bind to specific group of mRNAs encoding functionally related proteins.
| Target | RNP Complex |
|---|---|
| Product Type | Kit |
| Research Area | Epigenetics |
| Storage Temperature | 4°C |
| Protocols | |
| Regulatory Statement | For Research Use Only. Not for use in diagnostic procedures. |